Foreword¶
We have seen the use of jupyter notebooks.
As a matter of fact these slides are a notebook converted to (HTML) slides via the jupyter-nbconvert utility.
Get the code from gitlab here: https://gitlab.com/carlomt/pycourse with, in an new terminal:
git clone https://gitlab.com/carlomt/pycourse
cd pycourse
conda env create -f environment.yml
conda activate pycourse
. post-install.sh #Configure some extra stuff
The pre-installed environment course is enough to read/edit these slides.
Read the README.md file for additional instructions.
Python Introduction Course: Data Science tools¶
With emphasis on data-science problems
This course is available on gitlab
Contact us: andrea.dotti@gmail.com, mancinit@infn.it
The SciPy library contains several packages to perform specialized scientific calculations:
- Special functions (
scipy.special) - Integration (
scipy.integrate) - Optimization (
scipy.optimize) - Interpolation (
scipy.interpolate) - Fourier Transforms (
scipy.fftpack) - Signal Processing (
scipy.signal) - Linear Algebra (
scipy.linalg) - Sparse Eigenvalue Problems with ARPACK
- Compressed Sparse Graph Routines (
scipy.sparse.csgraph) - Spatial data structures and algorithms (
scipy.spatial) - Statistics (
scipy.stats) - Multidimensional image processing (
scipy.ndimage) - File IO (
scipy.io)
Numpy¶
It is the foundation of python scientific stack.
The basic building block is the numpy.array data structure. It can be used as a python list of numbers, but it is a specialized efficient way of manipulating numbers in python.
import numpy as np
a = np.array([1, 2, 3, 4], dtype=float)
a
array([1., 2., 3., 4.])
a = range(1000)
%timeit [ i**2 for i in a]
222 µs ± 3.93 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
b = np.arange(1000)
%timeit b**2
1.61 µs ± 10.6 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)
c = np.array([[1,2],[3,4]])
c
array([[1, 2],
[3, 4]])
c.ndim
2
c.shape
(2, 2)
c = np.arange(27)
c.reshape((3,3,3))
array([[[ 0, 1, 2],
[ 3, 4, 5],
[ 6, 7, 8]],
[[ 9, 10, 11],
[12, 13, 14],
[15, 16, 17]],
[[18, 19, 20],
[21, 22, 23],
[24, 25, 26]]])
np.zeros((2,2))
array([[0., 0.],
[0., 0.]])
np.ones((2,1))
array([[1.],
[1.]])
a = np.arange(27).reshape((3,3,3))
np.ones_like(a)
array([[[1, 1, 1],
[1, 1, 1],
[1, 1, 1]],
[[1, 1, 1],
[1, 1, 1],
[1, 1, 1]],
[[1, 1, 1],
[1, 1, 1],
[1, 1, 1]]])
np.eye(3)
array([[1., 0., 0.],
[0., 1., 0.],
[0., 0., 1.]])
a = np.arange(10)
a
array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
a[0]
0
a[-1]
9
a[0:3]
array([0, 1, 2])
a[::2]
array([0, 2, 4, 6, 8])
a = a.reshape(5,2)
a
array([[0, 1],
[2, 3],
[4, 5],
[6, 7],
[8, 9]])
a[3,1]
7
a[2,:]
array([4, 5])
Important: Views or copy¶
A slice or reshape is a view, simply a re-organization of the same data in memory, thus changing one element changes the same element in all views
a = np.arange(9)
b = a.reshape((3,3))
np.may_share_memory(a,b)
True
a[3] = -1
b
array([[ 0, 1, 2],
[-1, 4, 5],
[ 6, 7, 8]])
b = a.copy()
np.may_share_memory(a,b)
False
Boolean masks for extracting values¶
A typical operation done in your daily physics data analysis is to extract from an array the values that match a condition. Consider an array of the energies of particles, and assume you want to use only the energies above a given threshold. Boolean masking comes at a rescue
ene = np.random.exponential(size=10, scale=10.) # 1/scale e^(-ene/scale)
ene
array([ 4.85456007, 0.51860863, 3.69385318, 5.40459025, 3.83217684,
5.31482774, 0.20313695, 7.66005641, 2.52100998, 12.84343854])
mask = ene > 2
mask
array([ True, False, True, True, True, True, False, True, True,
True])
ene[mask]
array([ 4.85456007, 3.69385318, 5.40459025, 3.83217684, 5.31482774,
7.66005641, 2.52100998, 12.84343854])
ene[ene<2]
array([0.51860863, 0.20313695])
ene[ene<2] = 0
ene
array([ 4.85456007, 0. , 3.69385318, 5.40459025, 3.83217684,
5.31482774, 0. , 7.66005641, 2.52100998, 12.84343854])
Values with list of indexes¶
Similarly to boolean masks it is possible to access and modify values directly to an array using a list of indexes
status = np.random.randint(low=0,high=10,size=10)
status
array([2, 7, 7, 7, 8, 6, 0, 7, 4, 4])
status[[0, 3, 5]]
array([2, 7, 6])
status[[0, 3, 5]] = -1
status
array([-1, 7, 7, -1, 8, -1, 0, 7, 4, 4])
Simple Operations¶
a = np.arange(4)
a
array([0, 1, 2, 3])
a+1
array([1, 2, 3, 4])
10**a
array([ 1, 10, 100, 1000])
np.sin(a)
array([0. , 0.84147098, 0.90929743, 0.14112001])
Reductions¶
a = np.random.randint(low=0,high=10,size=4)
a
array([1, 8, 0, 9])
np.sum(a)
18
np.max(a), np.min(a)
(9, 0)
np.argmax(a), np.argmin(a)
(3, 2)
np.mean(a), np.median(a), np.std(a)
(4.5, 4.5, 4.031128874149275)
a = a.reshape(2,2)
a
array([[1, 8],
[0, 9]])
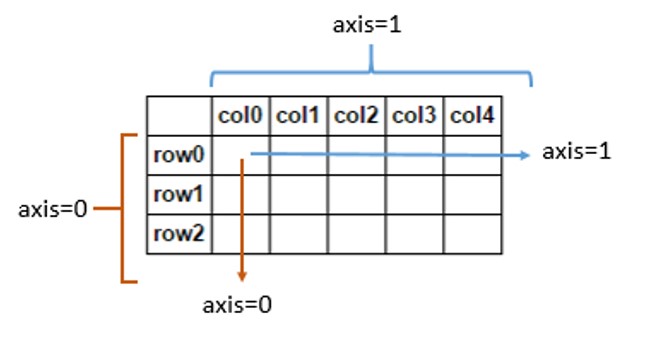
np.sum(a,axis=0)
array([ 1, 17])
m1 = a>0
m1
array([[ True, True],
[False, True]])
np.all(m1)
False
np.any(m1)
True
Matplotlib¶
Matplotlib is probably the most used Python package for 2D-graphics. It provides both a quick way to visualize data from Python and publication-quality figures in many formats.
Other visualization packages exists, often these are built on top of matplotlib.
The package is well integrated into IPython and Jupyter.
%matplotlib inline
from matplotlib import pyplot as plt
import numpy as np
X = np.linspace(-np.pi, np.pi, 256, endpoint=True)
C, S = np.cos(X), np.sin(X)
plt.plot(X,C)
plt.plot(X,S)
[<matplotlib.lines.Line2D at 0x7f6c7a21c4e0>]
Customization¶
plt.figure(figsize=(4, 3), dpi=80)
plt.plot(X, C, color="blue", linewidth=1.0, linestyle="-", label="cos")
plt.plot(X, S, color="green", linewidth=1.0, linestyle="-", label="sin")
plt.xlim(-4.0, 4.0)
plt.xticks(np.linspace(-4, 4, 9, endpoint=True))
plt.savefig("example.png", dpi=72)
plt.grid()
plt.xlabel("x")
plt.ylabel("y")
plt.title("Example")
plt.legend(loc="best")
<matplotlib.legend.Legend at 0x7f6c79c0bc18>
Multiple plots¶
plt.figure(figsize=(6, 4))
plt.subplot(2, 2, 1)
plt.plot(X, C, color="blue", linewidth=1.0, linestyle="-", label="cos")
plt.subplot(2, 2, 2)
plt.plot(X, S, color="green", linewidth=1.0, linestyle="-", label="sin")
plt.subplot(2, 2, 3)
plt.plot(X, C, color="red", linewidth=1.0, linestyle="-", label="cos")
plt.subplot(2, 2, 4)
plt.plot(X, S, color="black", linewidth=1.0, linestyle="-", label="sin")
plt.show()
Examples¶
plt.rcdefaults()
fig, ax = plt.subplots(figsize=(4,3))
# Example data
people = ('Tom', 'Dick', 'Harry', 'Slim', 'Jim')
y_pos = np.arange(len(people))
performance = 3 + 10 * np.random.rand(len(people))
error = np.random.rand(len(people))
ax.barh(y_pos, performance, xerr=error, align='center',
color='green', ecolor='black')
ax.set_yticks(y_pos)
ax.set_yticklabels(people)
ax.invert_yaxis() # labels read top-to-bottom
ax.set_xlabel('Performance')
ax.set_title('How fast do you want to go today?')
plt.show()
x = np.linspace(0, 1, 500)
y = np.sin(4 * np.pi * x) * np.exp(-5 * x)
fig, ax = plt.subplots()
ax.fill(x, y, zorder=10)
ax.grid(True, zorder=5)
plt.show()
fig, ax = plt.subplots()
for color in ['red', 'green', 'blue']:
n = 750
x, y = np.random.rand(2, n)
scale = 200.0 * np.random.rand(n)
ax.scatter(x, y, c=color, s=scale, label=color,
alpha=0.3, edgecolors='none')
ax.legend()
ax.grid(True)
plt.show()
mu = 200
sigma = 25
x = np.random.normal(mu, sigma, size=100)
fig, (ax0, ax1) = plt.subplots(ncols=2, figsize=(6, 3))
ax0.hist(x, 20, density=1, histtype='stepfilled', facecolor='g', alpha=0.75)
ax0.set_title('stepfilled')
# Create a histogram by providing the bin edges (unequally spaced).
bins = [100, 150, 180, 195, 205, 220, 250, 300]
ax1.hist(x, bins, density=1, histtype='bar', rwidth=0.8)
ax1.set_title('unequal bins')
plt.title(r'Histogram of IQ: $\mu=100$, $\sigma=15$');
from matplotlib import colors, ticker, cm
from scipy.stats import multivariate_normal
N = 100
x = np.linspace(-3.0, 3.0, N)
y = np.linspace(-2.0, 2.0, N)
X, Y = np.meshgrid(x, y)
pos = np.empty(X.shape+(2,))
pos[:,:,0] = X; pos[:,:,1] = Y
# A low hump with a spike coming out of the top right.
# Needs to have z/colour axis on a log scale so we see both hump and spike.
# linear scale only shows the spike.
z = (multivariate_normal([0.1, 0.2], [[1.0, 0.],[0, 1.0]]).pdf(pos)
+ 0.1 * (multivariate_normal([1.0, 1.0],[[0.01, 0.],[0., 0.01]])).pdf(pos))
# Automatic selection of levels works; setting the
# log locator tells contourf to use a log scale:
fig, ax = plt.subplots(figsize=(4,3))
cs = ax.contourf(X, Y, z, locator=ticker.LogLocator(), cmap=cm.PuBu_r)
cbar = fig.colorbar(cs)


Pandas¶
Pandas is a high-performance, high-level library that provides tools for data analysis.
It relies on the concept of DataFrame: a structured collection of data organized in records. This is the same concept of ROOT's NTuple that you are familiar with.
I think the name comes from R.
import numpy as np
import pandas as pd
s = pd.Series( [1., 2., 3., np.nan, 5. ], index=["a","b","c","d","e"])
s
a 1.0 b 2.0 c 3.0 d NaN e 5.0 dtype: float64
df = pd.DataFrame(
{
'Col1': [1.,2.,3.,4.],
'Col2': ["a","b","c","d"],
'Col3': [True, False, True, True]
}
)
df
| Col1 | Col2 | Col3 | |
|---|---|---|---|
| 0 | 1.0 | a | True |
| 1 | 2.0 | b | False |
| 2 | 3.0 | c | True |
| 3 | 4.0 | d | True |
Reading/Saving dataframes¶
Pandas support reading writing to several data formats, via specialized routines, many other formats, because dataframe (with other names) are a common concept:
| Format Type | Data Description | Reader | Writer |
|---|---|---|---|
| text | CSV | read_csv |
to_csv |
| text | JSON | read_json |
to_json |
| text | HTML | read_html |
to_html |
| text | Local clipboard | read_clipboard |
to_clipboard |
| binary | MS Excel | read_excel |
to_excel |
| binary | HDF5 Format | read_hdf |
to_hdf |
| binary | Feather Format | read_feather |
to_feather |
| binary | Parquet Format | read_parquet |
to_parquet |
| binary | Msgpack | read_msgpack |
to_msgpack |
| binary | Stata | read_stata |
to_stata |
| binary | SAS | read_sas |
|
| binary | Pickle Format | read_pickle |
to_pickle |
| SQL | SQL | read_sql |
to_sql |
| SQL | Google Big Query | read_gbq |
to_gbq |
As you can see the physicists ROOT format is not natively supported. However some external software to read TTrees are available. For example root_numpy, root_pandas, or uproot. ROOT usually comes with pre-installed pyROOT library (the one on the provided VM works only for python2), that offers basic functionalities.
df.dtypes
Col1 float64 Col2 object Col3 bool dtype: object
df.columns
Index(['Col1', 'Col2', 'Col3'], dtype='object')
df.index
RangeIndex(start=0, stop=4, step=1)
View data¶
df = pd.DataFrame( {'A':np.random.randint(0,10,100), 'B': [2**x for x in np.arange(100)], 'C':"a"})
df.head()
| A | B | C | |
|---|---|---|---|
| 0 | 4 | 1 | a |
| 1 | 2 | 2 | a |
| 2 | 0 | 4 | a |
| 3 | 9 | 8 | a |
| 4 | 3 | 16 | a |
df.tail(2)
| A | B | C | |
|---|---|---|---|
| 98 | 5 | 0 | a |
| 99 | 9 | 0 | a |
df.describe()
| A | B | |
|---|---|---|
| count | 100.000000 | 1.000000e+02 |
| mean | 4.420000 | -2.560000e+00 |
| std | 2.985419 | 1.070389e+18 |
| min | 0.000000 | -9.223372e+18 |
| 25% | 2.000000 | 0.000000e+00 |
| 50% | 5.000000 | 6.144000e+03 |
| 75% | 7.000000 | 1.717987e+11 |
| max | 9.000000 | 4.611686e+18 |
Select data¶
dates = pd.date_range('20190527',periods=7)
df = pd.DataFrame( np.random.rand(7,4), index=dates, columns=['A','B','C','D'])
df
| A | B | C | D | |
|---|---|---|---|---|
| 2019-05-27 | 0.944420 | 0.075201 | 0.167932 | 0.017186 |
| 2019-05-28 | 0.245307 | 0.577804 | 0.132167 | 0.372844 |
| 2019-05-29 | 0.459021 | 0.087459 | 0.647909 | 0.963480 |
| 2019-05-30 | 0.244232 | 0.261606 | 0.109693 | 0.494399 |
| 2019-05-31 | 0.575183 | 0.584652 | 0.113913 | 0.117457 |
| 2019-06-01 | 0.190005 | 0.692712 | 0.404453 | 0.995082 |
| 2019-06-02 | 0.931300 | 0.489561 | 0.193387 | 0.327648 |
df['A'] # or df.A
2019-05-27 0.632271 2019-05-28 0.691208 2019-05-29 0.603331 2019-05-30 0.043723 2019-05-31 0.552101 2019-06-01 0.330455 2019-06-02 0.841736 Freq: D, Name: A, dtype: float64
df[0:2]
| A | B | C | D | |
|---|---|---|---|---|
| 2019-05-27 | 0.632271 | 0.218192 | 0.139538 | 0.082094 |
| 2019-05-28 | 0.691208 | 0.700231 | 0.730246 | 0.328330 |
df['20190529':'20190531']
| A | B | C | D | |
|---|---|---|---|---|
| 2019-05-29 | 0.603331 | 0.165942 | 0.199283 | 0.119786 |
| 2019-05-30 | 0.043723 | 0.162669 | 0.291625 | 0.120803 |
| 2019-05-31 | 0.552101 | 0.102359 | 0.702799 | 0.455912 |
dates
DatetimeIndex(['2019-05-27', '2019-05-28', '2019-05-29', '2019-05-30',
'2019-05-31', '2019-06-01', '2019-06-02'],
dtype='datetime64[ns]', freq='D')
df.loc[dates[2]]
A 0.603331 B 0.165942 C 0.199283 D 0.119786 Name: 2019-05-29 00:00:00, dtype: float64
df.loc[dates[2],['B','C']]
B 0.165942 C 0.199283 Name: 2019-05-29 00:00:00, dtype: float64
df.iloc[2,2:4]
C 0.199283 D 0.119786 Name: 2019-05-29 00:00:00, dtype: float64
df[ df>0.5 ]
| A | B | C | D | |
|---|---|---|---|---|
| 2019-05-27 | 0.632271 | NaN | NaN | NaN |
| 2019-05-28 | 0.691208 | 0.700231 | 0.730246 | NaN |
| 2019-05-29 | 0.603331 | NaN | NaN | NaN |
| 2019-05-30 | NaN | NaN | NaN | NaN |
| 2019-05-31 | 0.552101 | NaN | 0.702799 | NaN |
| 2019-06-01 | NaN | NaN | 0.868320 | 0.510706 |
| 2019-06-02 | 0.841736 | NaN | 0.931434 | NaN |
Setting values¶
s = pd.Series( np.random.rand(7), index=dates )
s
2019-05-27 0.547899 2019-05-28 0.465972 2019-05-29 0.127552 2019-05-30 0.996297 2019-05-31 0.691513 2019-06-01 0.029567 2019-06-02 0.376882 Freq: D, dtype: float64
df['E'] = s
df
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.632271 | 0.218192 | 0.139538 | 0.082094 | 0.547899 |
| 2019-05-28 | 0.691208 | 0.700231 | 0.730246 | 0.328330 | 0.465972 |
| 2019-05-29 | 0.603331 | 0.165942 | 0.199283 | 0.119786 | 0.127552 |
| 2019-05-30 | 0.043723 | 0.162669 | 0.291625 | 0.120803 | 0.996297 |
| 2019-05-31 | 0.552101 | 0.102359 | 0.702799 | 0.455912 | 0.691513 |
| 2019-06-01 | 0.330455 | 0.227942 | 0.868320 | 0.510706 | 0.029567 |
| 2019-06-02 | 0.841736 | 0.088304 | 0.931434 | 0.300959 | 0.376882 |
df.loc[:,['C']] = 0
df
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.632271 | 0.218192 | 0.0 | 0.082094 | 0.547899 |
| 2019-05-28 | 0.691208 | 0.700231 | 0.0 | 0.328330 | 0.465972 |
| 2019-05-29 | 0.603331 | 0.165942 | 0.0 | 0.119786 | 0.127552 |
| 2019-05-30 | 0.043723 | 0.162669 | 0.0 | 0.120803 | 0.996297 |
| 2019-05-31 | 0.552101 | 0.102359 | 0.0 | 0.455912 | 0.691513 |
| 2019-06-01 | 0.330455 | 0.227942 | 0.0 | 0.510706 | 0.029567 |
| 2019-06-02 | 0.841736 | 0.088304 | 0.0 | 0.300959 | 0.376882 |
Operations¶
df.mean()
A 0.527832 B 0.237948 C 0.000000 D 0.274084 E 0.462240 dtype: float64
df.mean(axis=1)
2019-05-27 0.296091 2019-05-28 0.437148 2019-05-29 0.203322 2019-05-30 0.264698 2019-05-31 0.360377 2019-06-01 0.219734 2019-06-02 0.321576 Freq: D, dtype: float64
Merging dataframes¶
df1 = pd.DataFrame( np.random.rand(7,2), index=dates, columns=['A','B'])
df2 = pd.DataFrame( np.random.rand(7,3), index=dates, columns=['C','D','E'])
pd.concat([df1,df2],sort=False)
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.012169 | 0.252402 | NaN | NaN | NaN |
| 2019-05-28 | 0.420066 | 0.163832 | NaN | NaN | NaN |
| 2019-05-29 | 0.709032 | 0.134392 | NaN | NaN | NaN |
| 2019-05-30 | 0.245606 | 0.952309 | NaN | NaN | NaN |
| 2019-05-31 | 0.750060 | 0.851338 | NaN | NaN | NaN |
| 2019-06-01 | 0.334091 | 0.825410 | NaN | NaN | NaN |
| 2019-06-02 | 0.222300 | 0.897779 | NaN | NaN | NaN |
| 2019-05-27 | NaN | NaN | 0.781323 | 0.624619 | 0.382809 |
| 2019-05-28 | NaN | NaN | 0.932316 | 0.051429 | 0.823951 |
| 2019-05-29 | NaN | NaN | 0.246817 | 0.021852 | 0.699723 |
| 2019-05-30 | NaN | NaN | 0.700137 | 0.231148 | 0.373396 |
| 2019-05-31 | NaN | NaN | 0.340692 | 0.371376 | 0.751349 |
| 2019-06-01 | NaN | NaN | 0.567121 | 0.771248 | 0.712765 |
| 2019-06-02 | NaN | NaN | 0.970691 | 0.146501 | 0.218353 |
pd.concat([df1,df2],axis=1,join='inner')
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.012169 | 0.252402 | 0.781323 | 0.624619 | 0.382809 |
| 2019-05-28 | 0.420066 | 0.163832 | 0.932316 | 0.051429 | 0.823951 |
| 2019-05-29 | 0.709032 | 0.134392 | 0.246817 | 0.021852 | 0.699723 |
| 2019-05-30 | 0.245606 | 0.952309 | 0.700137 | 0.231148 | 0.373396 |
| 2019-05-31 | 0.750060 | 0.851338 | 0.340692 | 0.371376 | 0.751349 |
| 2019-06-01 | 0.334091 | 0.825410 | 0.567121 | 0.771248 | 0.712765 |
| 2019-06-02 | 0.222300 | 0.897779 | 0.970691 | 0.146501 | 0.218353 |
Grouping¶
s = pd.Series( ["a","b","a","c","a","c","b"], index=dates)
df['E']=s
df
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.632271 | 0.218192 | 0.0 | 0.082094 | a |
| 2019-05-28 | 0.691208 | 0.700231 | 0.0 | 0.328330 | b |
| 2019-05-29 | 0.603331 | 0.165942 | 0.0 | 0.119786 | a |
| 2019-05-30 | 0.043723 | 0.162669 | 0.0 | 0.120803 | c |
| 2019-05-31 | 0.552101 | 0.102359 | 0.0 | 0.455912 | a |
| 2019-06-01 | 0.330455 | 0.227942 | 0.0 | 0.510706 | c |
| 2019-06-02 | 0.841736 | 0.088304 | 0.0 | 0.300959 | b |
df.groupby('E').sum()
| A | B | C | D | |
|---|---|---|---|---|
| E | ||||
| a | 1.787704 | 0.486493 | 0.0 | 0.657791 |
| b | 1.532944 | 0.788535 | 0.0 | 0.629290 |
| c | 0.374178 | 0.390611 | 0.0 | 0.631508 |
Pivot table¶
dates = pd.date_range('20190527',periods=6, name='date')
df = pd.DataFrame( np.random.rand(6,3), index=dates, columns=['A','B','C'])
df['D'] = pd.Series(["a","a","b","b","c","c"],index=dates)
df['E'] = pd.Series(["one","two","one","two","one","two"],index=dates)
df
| A | B | C | D | E | |
|---|---|---|---|---|---|
| date | |||||
| 2019-05-27 | 0.474915 | 0.196110 | 0.301641 | a | one |
| 2019-05-28 | 0.057920 | 0.349846 | 0.670458 | a | two |
| 2019-05-29 | 0.878578 | 0.884794 | 0.904183 | b | one |
| 2019-05-30 | 0.456244 | 0.917951 | 0.879430 | b | two |
| 2019-05-31 | 0.592064 | 0.925768 | 0.601040 | c | one |
| 2019-06-01 | 0.403849 | 0.445142 | 0.004900 | c | two |
pd.pivot_table(df, values=['A','B','C'], index=['D','E'])
| A | B | C | ||
|---|---|---|---|---|
| D | E | |||
| a | one | 0.474915 | 0.196110 | 0.301641 |
| two | 0.057920 | 0.349846 | 0.670458 | |
| b | one | 0.878578 | 0.884794 | 0.904183 |
| two | 0.456244 | 0.917951 | 0.879430 | |
| c | one | 0.592064 | 0.925768 | 0.601040 |
| two | 0.403849 | 0.445142 | 0.004900 |
Plotting data¶
%matplotlib inline
df.plot()
<matplotlib.axes._subplots.AxesSubplot at 0x7f08ba132a20>
