Exercise solution¶
In [1]:
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
rng = np.random.default_rng(1234)
N = 3000
In [2]:
xy = rng.uniform(low=0., high=1., size=[2,N])
v = np.sum(xy**2, axis=0)
In [3]:
w, h = plt.figaspect(1.)
plt.figure(figsize=(w,h))
plt.grid(True)
plt.xlabel('x',labelpad=0.5)
plt.ylabel('y',labelpad=0.5)
plt.scatter(xy[0,:], xy[1,:], s=2)
plt.scatter(xy[0,:][v<1], xy[1,:][v<1], s=2)
Out[3]:
<matplotlib.collections.PathCollection at 0x119d54820>
In [4]:
values = np.cumsum(np.sum(rng.uniform(0,1,size=[2,N])**2,axis=0)<1) \
/(np.arange(1,N+1))*4
In [5]:
plt.grid(True)
plt.xlabel('n',labelpad=0.5)
plt.ylabel('n/N*4',labelpad=0.5)
plt.plot(np.arange(1,N+1), np.ones(N)*np.pi)
plt.plot(np.arange(1,N+1), values)
plt.yticks([np.pi/2, np.pi, np.pi*1.5], [u'\u03c0/2',u'\u03c0',u'\u03c0*3/2'] )
plt.show()
you can do the plot in logscale and with $\frac{1}{\sqrt(N)}$
In [6]:
plt.grid(True)
plt.xlabel('n',labelpad=0.5)
plt.ylabel('n/N*4',labelpad=0.5)
plt.plot(np.arange(1,N+1), 1/np.sqrt(np.arange(1,N+1))+np.pi)
plt.plot(np.arange(1,N+1), np.ones(N)*np.pi)
plt.plot(np.arange(1,N+1), values)
plt.yticks([np.pi/2, np.pi, np.pi*1.5], [u'\u03c0/2',u'\u03c0',u'\u03c0*3/2'] )
plt.yscale('log')
plt.show()
Pandas¶
Pandas is a high-performance, high-level library that provides tools for data analysis.
It relies on the concept of DataFrame: a structured collection of data organized in records. This is the same concept of ROOT's NTuple that you are familiar with.
I think the name comes from R.
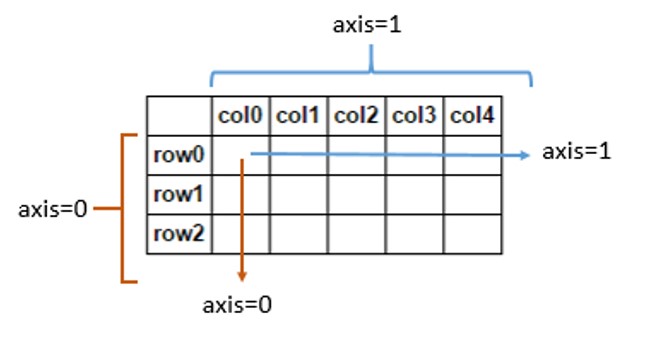
In [7]:
import numpy as np
import pandas as pd
s = pd.Series( [1., 2., 3., np.nan, 5. ], index=["a","b","c","d","e"])
s
Out[7]:
a 1.0 b 2.0 c 3.0 d NaN e 5.0 dtype: float64
In [8]:
df = pd.DataFrame(
{
'Col1': [1.,2.,3.,4.],
'Col2': ["a","b","c","d"],
'Col3': [True, False, True, True]
}
)
df
Out[8]:
| Col1 | Col2 | Col3 | |
|---|---|---|---|
| 0 | 1.0 | a | True |
| 1 | 2.0 | b | False |
| 2 | 3.0 | c | True |
| 3 | 4.0 | d | True |
Reading/Saving dataframes¶
Pandas support reading writing to several data formats, via specialized routines, many other formats, because dataframe (with other names) are a common concept:
| Format Type | Data Description | Reader | Writer |
|---|---|---|---|
| text | CSV | read_csv |
to_csv |
| text | JSON | read_json |
to_json |
| text | HTML | read_html |
to_html |
| text | Local clipboard | read_clipboard |
to_clipboard |
| binary | MS Excel | read_excel |
to_excel |
| binary | HDF5 Format | read_hdf |
to_hdf |
| binary | Feather Format | read_feather |
to_feather |
| binary | Parquet Format | read_parquet |
to_parquet |
| binary | Msgpack | read_msgpack |
to_msgpack |
| binary | Stata | read_stata |
to_stata |
| binary | SAS | read_sas |
|
| binary | Pickle Format | read_pickle |
to_pickle |
| SQL | SQL | read_sql |
to_sql |
| SQL | Google Big Query | read_gbq |
to_gbq |
As you can see the physicists ROOT format is not natively supported. However some external software to read TTrees are available. For example uproot. ROOT usually comes with pre-installed pyROOT library.
In [9]:
df.dtypes
Out[9]:
Col1 float64 Col2 object Col3 bool dtype: object
In [10]:
df.columns
Out[10]:
Index(['Col1', 'Col2', 'Col3'], dtype='object')
In [11]:
df.index
Out[11]:
RangeIndex(start=0, stop=4, step=1)
View data¶
In [12]:
df = pd.DataFrame( {'A':np.random.randint(0,10,100), 'B': [2**x for x in np.arange(100)], 'C':"a"})
df.head()
Out[12]:
| A | B | C | |
|---|---|---|---|
| 0 | 0 | 1 | a |
| 1 | 6 | 2 | a |
| 2 | 4 | 4 | a |
| 3 | 5 | 8 | a |
| 4 | 2 | 16 | a |
In [13]:
df.tail(2)
Out[13]:
| A | B | C | |
|---|---|---|---|
| 98 | 7 | 0 | a |
| 99 | 9 | 0 | a |
In [14]:
df.describe()
Out[14]:
| A | B | |
|---|---|---|
| count | 100.000000 | 1.000000e+02 |
| mean | 4.850000 | -2.560000e+00 |
| std | 2.793842 | 1.070389e+18 |
| min | 0.000000 | -9.223372e+18 |
| 25% | 3.000000 | 0.000000e+00 |
| 50% | 5.000000 | 6.144000e+03 |
| 75% | 7.000000 | 1.717987e+11 |
| max | 9.000000 | 4.611686e+18 |
Select data¶
In [15]:
dates = pd.date_range('20190527',periods=7)
df = pd.DataFrame( np.random.rand(7,4), index=dates, columns=['A','B','C','D'])
df
Out[15]:
| A | B | C | D | |
|---|---|---|---|---|
| 2019-05-27 | 0.422593 | 0.039869 | 0.363634 | 0.827147 |
| 2019-05-28 | 0.981383 | 0.226043 | 0.943508 | 0.716723 |
| 2019-05-29 | 0.154992 | 0.660813 | 0.407191 | 0.779359 |
| 2019-05-30 | 0.474430 | 0.753383 | 0.428391 | 0.253242 |
| 2019-05-31 | 0.334128 | 0.438464 | 0.995652 | 0.979889 |
| 2019-06-01 | 0.616849 | 0.192285 | 0.637097 | 0.687652 |
| 2019-06-02 | 0.572270 | 0.989857 | 0.226496 | 0.511833 |
In [16]:
df['A'] # or df.A
Out[16]:
2019-05-27 0.422593 2019-05-28 0.981383 2019-05-29 0.154992 2019-05-30 0.474430 2019-05-31 0.334128 2019-06-01 0.616849 2019-06-02 0.572270 Freq: D, Name: A, dtype: float64
In [17]:
df[0:2]
Out[17]:
| A | B | C | D | |
|---|---|---|---|---|
| 2019-05-27 | 0.422593 | 0.039869 | 0.363634 | 0.827147 |
| 2019-05-28 | 0.981383 | 0.226043 | 0.943508 | 0.716723 |
In [18]:
df['20190529':'20190531']
Out[18]:
| A | B | C | D | |
|---|---|---|---|---|
| 2019-05-29 | 0.154992 | 0.660813 | 0.407191 | 0.779359 |
| 2019-05-30 | 0.474430 | 0.753383 | 0.428391 | 0.253242 |
| 2019-05-31 | 0.334128 | 0.438464 | 0.995652 | 0.979889 |
In [19]:
dates
Out[19]:
DatetimeIndex(['2019-05-27', '2019-05-28', '2019-05-29', '2019-05-30',
'2019-05-31', '2019-06-01', '2019-06-02'],
dtype='datetime64[ns]', freq='D')
In [20]:
df.loc[dates[2]]
Out[20]:
A 0.154992 B 0.660813 C 0.407191 D 0.779359 Name: 2019-05-29 00:00:00, dtype: float64
In [21]:
df.loc[dates[2],['B','C']]
Out[21]:
B 0.660813 C 0.407191 Name: 2019-05-29 00:00:00, dtype: float64
In [22]:
df.iloc[2,1:3]
Out[22]:
B 0.660813 C 0.407191 Name: 2019-05-29 00:00:00, dtype: float64
In [23]:
df[ df>0.5 ]
Out[23]:
| A | B | C | D | |
|---|---|---|---|---|
| 2019-05-27 | NaN | NaN | NaN | 0.827147 |
| 2019-05-28 | 0.981383 | NaN | 0.943508 | 0.716723 |
| 2019-05-29 | NaN | 0.660813 | NaN | 0.779359 |
| 2019-05-30 | NaN | 0.753383 | NaN | NaN |
| 2019-05-31 | NaN | NaN | 0.995652 | 0.979889 |
| 2019-06-01 | 0.616849 | NaN | 0.637097 | 0.687652 |
| 2019-06-02 | 0.572270 | 0.989857 | NaN | 0.511833 |
Setting values¶
In [24]:
s = pd.Series( np.random.rand(7), index=dates )
s
Out[24]:
2019-05-27 0.078480 2019-05-28 0.572171 2019-05-29 0.448760 2019-05-30 0.697107 2019-05-31 0.044482 2019-06-01 0.310467 2019-06-02 0.440914 Freq: D, dtype: float64
In [25]:
df['E'] = s
df
Out[25]:
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.422593 | 0.039869 | 0.363634 | 0.827147 | 0.078480 |
| 2019-05-28 | 0.981383 | 0.226043 | 0.943508 | 0.716723 | 0.572171 |
| 2019-05-29 | 0.154992 | 0.660813 | 0.407191 | 0.779359 | 0.448760 |
| 2019-05-30 | 0.474430 | 0.753383 | 0.428391 | 0.253242 | 0.697107 |
| 2019-05-31 | 0.334128 | 0.438464 | 0.995652 | 0.979889 | 0.044482 |
| 2019-06-01 | 0.616849 | 0.192285 | 0.637097 | 0.687652 | 0.310467 |
| 2019-06-02 | 0.572270 | 0.989857 | 0.226496 | 0.511833 | 0.440914 |
In [26]:
df.loc[:,['C']] = 0
df
Out[26]:
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.422593 | 0.039869 | 0 | 0.827147 | 0.078480 |
| 2019-05-28 | 0.981383 | 0.226043 | 0 | 0.716723 | 0.572171 |
| 2019-05-29 | 0.154992 | 0.660813 | 0 | 0.779359 | 0.448760 |
| 2019-05-30 | 0.474430 | 0.753383 | 0 | 0.253242 | 0.697107 |
| 2019-05-31 | 0.334128 | 0.438464 | 0 | 0.979889 | 0.044482 |
| 2019-06-01 | 0.616849 | 0.192285 | 0 | 0.687652 | 0.310467 |
| 2019-06-02 | 0.572270 | 0.989857 | 0 | 0.511833 | 0.440914 |
Operations¶
In [27]:
df.mean()
Out[27]:
A 0.508092 B 0.471531 C 0.000000 D 0.679406 E 0.370340 dtype: float64
In [28]:
df.mean(axis=1)
Out[28]:
2019-05-27 0.273618 2019-05-28 0.499264 2019-05-29 0.408785 2019-05-30 0.435632 2019-05-31 0.359393 2019-06-01 0.361450 2019-06-02 0.502975 Freq: D, dtype: float64
Merging dataframes¶
In [29]:
df1 = pd.DataFrame( np.random.rand(7,2), index=dates, columns=['A','B'])
df2 = pd.DataFrame( np.random.rand(7,3), index=dates, columns=['C','D','E'])
pd.concat([df1,df2],sort=False)
Out[29]:
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.838082 | 0.415426 | NaN | NaN | NaN |
| 2019-05-28 | 0.787008 | 0.931342 | NaN | NaN | NaN |
| 2019-05-29 | 0.144897 | 0.392980 | NaN | NaN | NaN |
| 2019-05-30 | 0.156783 | 0.775479 | NaN | NaN | NaN |
| 2019-05-31 | 0.096234 | 0.405344 | NaN | NaN | NaN |
| 2019-06-01 | 0.568132 | 0.352329 | NaN | NaN | NaN |
| 2019-06-02 | 0.128113 | 0.230876 | NaN | NaN | NaN |
| 2019-05-27 | NaN | NaN | 0.404065 | 0.922900 | 0.860763 |
| 2019-05-28 | NaN | NaN | 0.548356 | 0.370591 | 0.767548 |
| 2019-05-29 | NaN | NaN | 0.053025 | 0.846114 | 0.898454 |
| 2019-05-30 | NaN | NaN | 0.877660 | 0.556573 | 0.621617 |
| 2019-05-31 | NaN | NaN | 0.063489 | 0.816351 | 0.059653 |
| 2019-06-01 | NaN | NaN | 0.648635 | 0.929256 | 0.322937 |
| 2019-06-02 | NaN | NaN | 0.803799 | 0.484108 | 0.862330 |
In [30]:
pd.concat([df1,df2],axis=1,join='inner') #same syntax as for db (ineer, outer, left, right)
Out[30]:
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.838082 | 0.415426 | 0.404065 | 0.922900 | 0.860763 |
| 2019-05-28 | 0.787008 | 0.931342 | 0.548356 | 0.370591 | 0.767548 |
| 2019-05-29 | 0.144897 | 0.392980 | 0.053025 | 0.846114 | 0.898454 |
| 2019-05-30 | 0.156783 | 0.775479 | 0.877660 | 0.556573 | 0.621617 |
| 2019-05-31 | 0.096234 | 0.405344 | 0.063489 | 0.816351 | 0.059653 |
| 2019-06-01 | 0.568132 | 0.352329 | 0.648635 | 0.929256 | 0.322937 |
| 2019-06-02 | 0.128113 | 0.230876 | 0.803799 | 0.484108 | 0.862330 |
Grouping¶
In [31]:
s = pd.Series( ["a","b","a","c","a","c","b"], index=dates)
df['E']=s
df
Out[31]:
| A | B | C | D | E | |
|---|---|---|---|---|---|
| 2019-05-27 | 0.422593 | 0.039869 | 0 | 0.827147 | a |
| 2019-05-28 | 0.981383 | 0.226043 | 0 | 0.716723 | b |
| 2019-05-29 | 0.154992 | 0.660813 | 0 | 0.779359 | a |
| 2019-05-30 | 0.474430 | 0.753383 | 0 | 0.253242 | c |
| 2019-05-31 | 0.334128 | 0.438464 | 0 | 0.979889 | a |
| 2019-06-01 | 0.616849 | 0.192285 | 0 | 0.687652 | c |
| 2019-06-02 | 0.572270 | 0.989857 | 0 | 0.511833 | b |
In [32]:
df.groupby('E').sum()
Out[32]:
| A | B | C | D | |
|---|---|---|---|---|
| E | ||||
| a | 0.911712 | 1.139146 | 0 | 2.586396 |
| b | 1.553653 | 1.215900 | 0 | 1.228555 |
| c | 1.091279 | 0.945668 | 0 | 0.940894 |
Pivot table¶
In [33]:
dates = pd.date_range('20190527',periods=6, name='date')
df = pd.DataFrame( np.random.rand(6,3), index=dates, columns=['A','B','C'])
df['D'] = pd.Series(["a","a","b","b","c","c"],index=dates)
df['E'] = pd.Series(["one","two","one","two","one","two"],index=dates)
df
Out[33]:
| A | B | C | D | E | |
|---|---|---|---|---|---|
| date | |||||
| 2019-05-27 | 0.646359 | 0.040318 | 0.706039 | a | one |
| 2019-05-28 | 0.983550 | 0.713946 | 0.066779 | a | two |
| 2019-05-29 | 0.883759 | 0.702453 | 0.889921 | b | one |
| 2019-05-30 | 0.378029 | 0.356373 | 0.709079 | b | two |
| 2019-05-31 | 0.227522 | 0.062629 | 0.230672 | c | one |
| 2019-06-01 | 0.024039 | 0.821423 | 0.490518 | c | two |
In [34]:
pd.pivot_table(df, values=['A','B','C'], index=['D','E'])
Out[34]:
| A | B | C | ||
|---|---|---|---|---|
| D | E | |||
| a | one | 0.646359 | 0.040318 | 0.706039 |
| two | 0.983550 | 0.713946 | 0.066779 | |
| b | one | 0.883759 | 0.702453 | 0.889921 |
| two | 0.378029 | 0.356373 | 0.709079 | |
| c | one | 0.227522 | 0.062629 | 0.230672 |
| two | 0.024039 | 0.821423 | 0.490518 |
Plotting data¶
In [35]:
df.plot()
Out[35]:
<AxesSubplot:xlabel='date'>