Using JAGS
Let us repeat the Monte Carlo simulation
improperly using the program JAGS [24],
interfaced to R via the package rjags [26].
JAGS is a powerful tool developed,
as open source, multi-platform clone of BUGS,40
to perform Bayesian inference
by Markov Chain Monte Carlo (MCMC)
using the Gibbs Sampler algorithm, as its name
reminds, acronym of Just Another Gibbs Sampler.
For the moment we just get familiar with JAGS using it as
kind of `curious' random generator.
The first thing to do is to write down
the probabilistic model that relates the different variables
that enter the game. For example, the
left hand graphical model of Fig. ![[*]](crossref.png) is implemented in JAGS by the
following self-explaining piece of code
is implemented in JAGS by the
following self-explaining piece of code
model {
n.I ~ dbin(p, ns)
n.NI <- ns - n.I
nP.I ~ dbin(pi1, n.I)
nP.NI ~ dbin(pi2, n.NI)
nP ~ sum(nP.I, nP.NI)
fP <- nP / ns
}
in which we have added the last instruction
to model the trivial node (not shown in Fig. ![[*]](crossref.png) )
relating in a deterministic way fP to nP and ns.
In the model the symbol `
)
relating in a deterministic way fP to nP and ns.
In the model the symbol `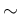 ' indicates that, e.g. n.I is described
by a binomial distribution defined by p and ns
(be aware of the different order of the parameters with respect to the
R function!), while `
' indicates that, e.g. n.I is described
by a binomial distribution defined by p and ns
(be aware of the different order of the parameters with respect to the
R function!), while ` '
stands for a deterministic relation
(indeed the symbols of assignment in R).
A nice thing of such a model is that the order of the instructions
is not relevant.
In fact it is only needed - let us put it so -
to describe the related graphical model. All the rest will
be done internally by JAGS at the compilation step.
'
stands for a deterministic relation
(indeed the symbols of assignment in R).
A nice thing of such a model is that the order of the instructions
is not relevant.
In fact it is only needed - let us put it so -
to describe the related graphical model. All the rest will
be done internally by JAGS at the compilation step.
Then, obviously, we have to
- pass to the program
the model parameters (observed nodes), which are
p, ns, pi1 and pi2;
- instruct it on how many `iterations' to do;
- analyze, among all unobserved nodes
(n.I, n.NI, nP.I, nP.NI, nP
and fP), the ones of interest, the most important one
being, for us, fP.
Moving to the second model of Fig. ![[*]](crossref.png) ,
in which we also take into account the uncertainty about
,
in which we also take into account the uncertainty about
 and
and  , is straightforward: we just need to add
two instructions to tell JAGS that
pi1 and pi2 are indeed unobserved
and that they depend on r1, s1, r2 and s2:
, is straightforward: we just need to add
two instructions to tell JAGS that
pi1 and pi2 are indeed unobserved
and that they depend on r1, s1, r2 and s2:
model {
n.I ~ dbin(p, ns)
n.NI <- ns - n.I
nP.I ~ dbin(pi1, n.I)
nP.NI ~ dbin(pi2, n.NI)
pi1 ~ dbeta(r1, s1)
pi2 ~ dbeta(r2, s2)
nP ~ sum(nP.I, nP.NI)
fP <- nP / ns
}
Once the model is defined, it has to be saved into a file,
whose location is then
passed to JAGS
(we shall regularly use the temporary file tmp_model.bug,
whose extension `.bug' is the BUGS/JAGS default).
At this point, moving to the R code to interact
with JAGS, we need to
- load the interfacing package rjags;
- prepare a R `list' containing the data (in particular, the values
of the `observed' nodes);
- call jags.model() to `setup' the model;
- call coda.samples() to ask JAGS to perform the sampling,
also specifying the variables we want to monitor,
whose `histories' will be returned in a single
`object'.41
Finally we have to show the result. All this is done, for example,
in the R script provided
in Appendix B.6 (note that the temporary model file is written
directly from R, a convenient solution for small models).
Needless to say, we get, apart from statistical fluctuations
inherent to
Monte Carlo methods, `exactly' the same results obtained with the
script of Appendix B.5, which only uses R statistical functions.
![[*]](crossref.png) is implemented in JAGS by the
following self-explaining piece of code
is implemented in JAGS by the
following self-explaining piece of code
![[*]](crossref.png) )
relating in a deterministic way fP to nP and ns.
In the model the symbol `
)
relating in a deterministic way fP to nP and ns.
In the model the symbol `![[*]](crossref.png) ,
in which we also take into account the uncertainty about
,
in which we also take into account the uncertainty about