Next: Sceptical combination with JAGS Up: Sceptical combination of experimental Previous: Implementation in JAGS/rjags
 |
For the parameters
of the Gamma pdf
of ![]() we stick to those chosen in Ref. [15],18
i.e.
we stick to those chosen in Ref. [15],18
i.e. ![]() and
and ![]() ,
in order to get
,
in order to get
![]() .
Figure 9,
.
Figure 9,
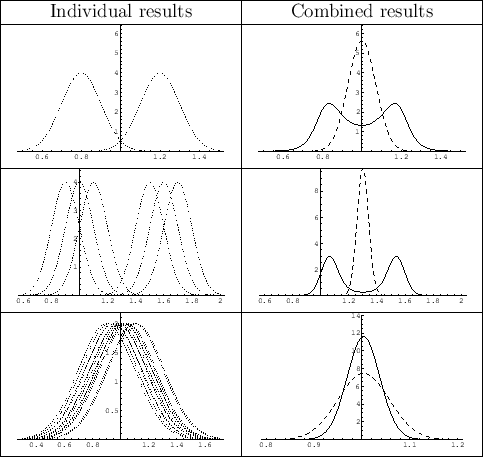 |
Particularly interesting are the cases in which
the individual results cluster in two regions, or when
they overlap `too much'. In the first case, while the simple
weighted average prefers mass values in a region where there is no experimental
support, the sceptical combination exhibits a bimodal distribution,
because we tend to believe with equal probability that the true value
is in either side
(but it could also be in the middle, although with
low probability). In the second case, instead, in which the results overlap
too much, the method has the nice feature of
producing a pdf narrower than that obtained by the
simple weighted average, reflecting our natural suspicion
that the quoted uncertainties might have been
overestimated. In Ref. [15] (Fig. 5 there)
it was also studied how the results varied if the initial
![]() was allowed to move by
was allowed to move by ![]() .
This leads us to be rather
confident that the choice of
.
This leads us to be rather
confident that the choice of ![]() and
and ![]() is not critical.
is not critical.
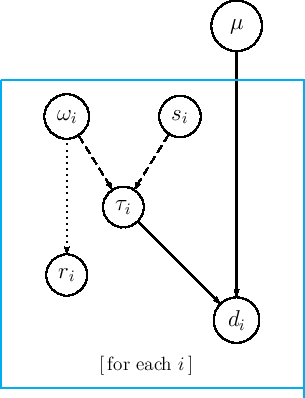
Here is, finally, the JAGS model, easy to understand if we compare it with the graphical model of Fig. 8:
var tau[N], r[N], omega[N];
model {
for (i in 1:N) {
d[i] ~ dnorm(mu, tau[i]);
tau[i] <- omega[i]/s[i]^2;
omega[i] ~ dgamma(delta, lambda);
r[i] <- 1.0/sqrt(omega[i]);
}
mu ~ dnorm(0.0, 1.0E-8);
}
Before running it, let us make a very trivial model
in which JAGS is used as a simple random generator, without
any inferential purpose, just to get confidence with the
prior distribution of ![]() . Moreover, consisting
the core of the model of just two lines of code,
we write it directly from the R script into a temporary file.
Here is the complete script, in which it also shown an
alternative way (indeed the simplest one in R)
to prepare the `list' data to be passed to JAGS
via jags.model() - note the missing update(), because
we deal here with direct sampling and there are no burn-in issues:
. Moreover, consisting
the core of the model of just two lines of code,
we write it directly from the R script into a temporary file.
Here is the complete script, in which it also shown an
alternative way (indeed the simplest one in R)
to prepare the `list' data to be passed to JAGS
via jags.model() - note the missing update(), because
we deal here with direct sampling and there are no burn-in issues:
library(rjags)
data = list(delta=1.3, lambda=0.6)
model = "tmp_model.bug"
write("
model{
omega ~ dgamma(delta, lambda);
r <- 1.0/sqrt(omega);
}
", model)
jm <- jags.model(model, data)
chain <- coda.samples(jm, c("omega","r"), n.iter=10000)
plot(chain)
print(summary(chain))
This is the result of the last command:
1. Empirical mean and standard deviation for each variable,
plus standard error of the mean:
Mean SD Naive SE Time-series SE
omega 2.1979 1.9127 0.019127 0.019065
r 0.9948 0.9096 0.009096 0.009096
2. Quantiles for each variable:
2.5% 25% 50% 75% 97.5%
omega 0.1162 0.8045 1.6582 3.035 7.329
r 0.3694 0.5741 0.7766 1.115 2.934
We see that the (indirectly) sampled r has
(with good approximation)
the expected unitary mean and standard deviation.
As a further check, let us use directly the Gamma random
generator rgamma() of R,
r <- 1/sqrt(rgamma(10000, 1.3, 0.6))
cat(sprintf("mean(r) = %f, sd(r) = %f\n", mean(r), sd(r)))
whose (aleatory) result is left as exercise to the reader.