Next: Efficacy against disease severity Up: What is the probability Previous: Taking into account `informative
![[*]](crossref.png) .
.
 |
![[*]](crossref.png) ,
and do the remaining work with `direct' Monte Carlo.
However, having observed that the resulting pdf of
,
and do the remaining work with `direct' Monte Carlo.
However, having observed that the resulting pdf of
mu <- 0.944; sigma <- 0.019; e.rep <- 0.950 # Pfizer
# mu <- 0.941; sigma <- 0.019; e.rep <- 0.941 # Moderna
# mu <- 0.861; sigma <- 0.075; e.rep <- 0.900 # AZ LDSD
# mu <- 0.599; sigma <- 0.090; e.rep <- 0.621 # AZ SDSD
# uncomment the following line to simulate a negligible uncertainty
# sigma <- 0.0001
r = (1-mu)*mu^2/sigma^2 - mu
s = r*(1-mu)/mu
cat(sprintf("r = %.2f, s = %.2f\n", r, s))
ns <- 1000000
nV <- 100000
pA <- 0.01
nA <- rbinom(ns, nV, pA)
cat(sprintf("nA: mean+-sigma: %.1f +- %.1f\n", mean(nA), sd(nA)))
eps <- rbeta(ns,r,s)
nvI <- rbinom(ns, nA, 1-eps)
hist(nvI, nc=100, col='cyan', freq=FALSE, main=”)
cat(sprintf("nvI: mean+-sigma: %.1f +- %.1f\n", mean(nvI), sd(nvI)))
lines(rep(pA*nV*(1-mu), 2), c(0,1), col='red', lty=1, lwd=2)
lines(rep(pA*nV*(1-e.rep), 2), c(0,1), col='red', lty=2, lwd=2)
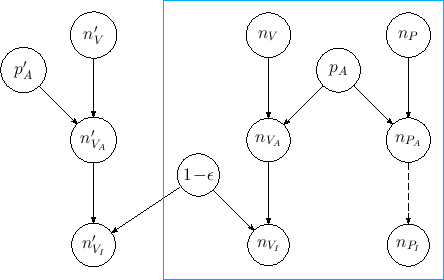
A number of hundred thousand vaccinated individuals
has been used, with
an absolutely hypothetical value of assault probability
of 1 %.
The script can also be used to simulate the effect of a precise
value of 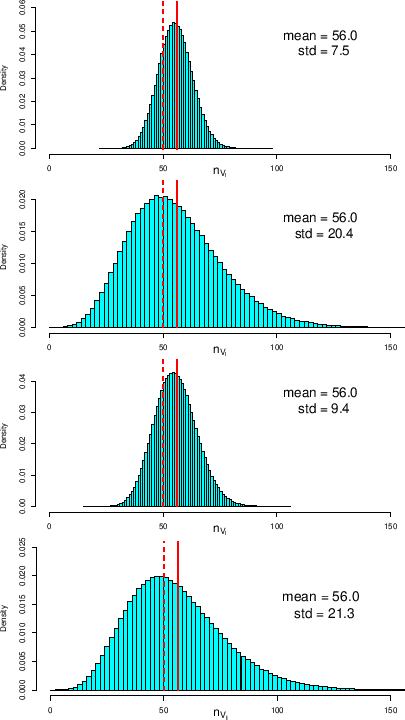 |
![[*]](crossref.png) .
In this idealized case the distribution of
.
In this idealized case the distribution of
| Binom |
![[*]](crossref.png) .
.
The effect of the uncertainty about ![]() is shown in the
second (top-down) histogram of the same figure. As we can see,
the distribution becomes remarkably wider and more asymmetric,
with a right-hand skewness, effect of the left-hand skewness of
is shown in the
second (top-down) histogram of the same figure. As we can see,
the distribution becomes remarkably wider and more asymmetric,
with a right-hand skewness, effect of the left-hand skewness of
![]() .
We see then, in the third histogram, the effect
of a hypothetical uncertainty about
.
We see then, in the third histogram, the effect
of a hypothetical uncertainty about ![]() , modeled here
with a standard deviation of
, modeled here
with a standard deviation of
![]() (but this has to be understood
really as an exercise done only to have an idea of the effect,
because a reasonable uncertainty could indeed be much larger).
Finally, including both sources of uncertainty, we get
the histogram and the numbers at the bottom of the figure.
Vertical lines show the predicted values for
(but this has to be understood
really as an exercise done only to have an idea of the effect,
because a reasonable uncertainty could indeed be much larger).
Finally, including both sources of uncertainty, we get
the histogram and the numbers at the bottom of the figure.
Vertical lines show the predicted values for ![]() by using the MCMC mean value (solid line) and using the
modal value (dashed lines).
by using the MCMC mean value (solid line) and using the
modal value (dashed lines).
As a further step, following Ref. [17]
(see in particular Secs. 5.2.1 and 5.3.1 there),
let us try to get approximated formulae for the expected value
and the standard deviation of ![]() . The idea, we shortly remind,
is to start with the expected value and variance evaluated
for the expected values of
. The idea, we shortly remind,
is to start with the expected value and variance evaluated
for the expected values of ![]() and
and ![]() , and then
make a `propagation of uncertainty' by linearization
(as if
, and then
make a `propagation of uncertainty' by linearization
(as if ![]() and
and ![]() were `systematics').
Here are the resulting formulae
were `systematics').
Here are the resulting formulae