Next: Comparison of the results Up: Use of MCMC methods Previous: Model A (Fig. ), with
![[*]](crossref.png) ), with flat priors
on
), with flat priors
on ![[*]](crossref.png) , whose
implementation in the JAGS language is the following:
, whose
implementation in the JAGS language is the following:
model {
x1 ~ dpois(lambda1)
x2 ~ dpois(lambda2)
lambda1 <- r1 * T1
lambda2 <- r2 * T2
r1 <- rho * r2
r2 ~ dgamma(1, 1e-6)
rho ~ dgamma(1, 1e-6)
}
The complete R script, which
uses the same data (
![[*]](crossref.png) and the details are given in the following printouts
and the details are given in the following printouts
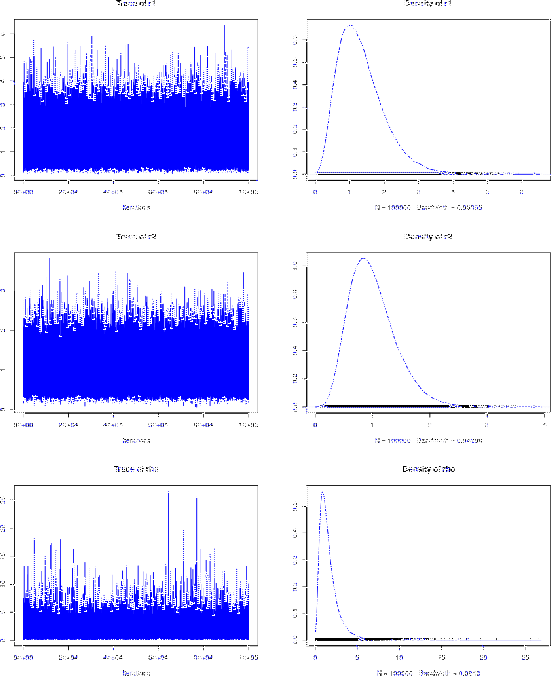 |
1. Empirical mean and standard deviation for each variable,
plus standard error of the mean:
Mean SD Naive SE Time-series SE
r1 1.334 0.6694 0.002117 0.002117
r2 1.002 0.4068 0.001286 0.001925
rho 1.595 1.1918 0.003769 0.006058
2. Quantiles for each variable:
2.5% 25% 50% 75% 97.5%
r1 0.3616 0.8438 1.2234 1.704 2.941
r2 0.3671 0.7061 0.9477 1.238 1.940
rho 0.3167 0.8199 1.2923 2.012 4.638
Exact:
r1 = 1.333 +- 0.667
r2 = 1.000 +- 0.408
rho = 1.600 +- 1.200
Again, the agreement between the MCMC and the exact results is excellent.