Next: Model B (Fig. ), with Up: Use of MCMC methods Previous: Use of MCMC methods
![[*]](crossref.png) ), with flat priors
on
), with flat priors
on ![[*]](crossref.png) .
The code that instructs JAGS about the model is
practically a transcription
of the expressions to state that a variable follows a given distribution.
Therefore, since we have
.
The code that instructs JAGS about the model is
practically a transcription
of the expressions to state that a variable follows a given distribution.
Therefore, since we have
| Poisson |
(125) | ||
| Poisson |
(126) | ||
| (127) | |||
| (128) | |||
| (129) |
model {
x1 ~ dpois(lambda1)
x2 ~ dpois(lambda2)
lambda1 <- r1 * T1
lambda2 <- r2 * T2
r1 ~ dgamma(1, 1e-6)
r2 ~ dgamma(1, 1e-6)
rho <- r1/r2
}
in which are also included the flat priors of
| Gamma |
(130) | ||
| Gamma |
(131) |
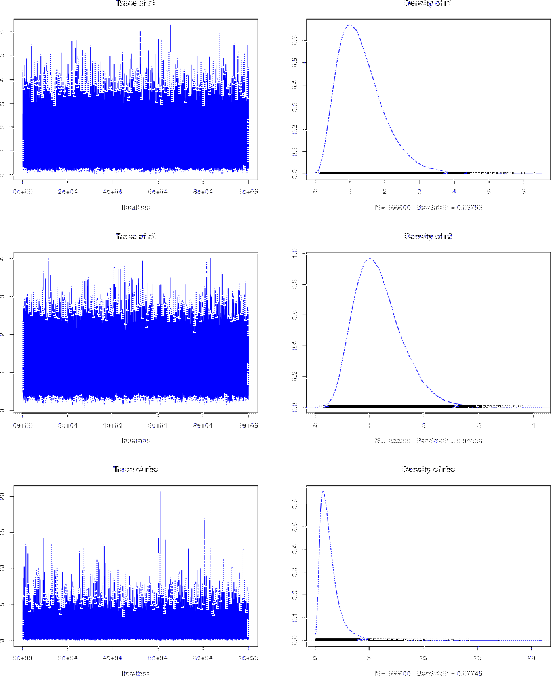 |
The summary figure ![[*]](crossref.png) , drawn
automatically by R when
the command plot() is called with first argument an MCMC
chain object, shows, for each of the three variables that we have
chosen to monitor,
the `trace' and the 'density'.
The latter is a smoothed representation of the histogram
of the possible occurrences of a variable in the chain.
The former shows the `history'
of a variable during the sampling, and it is important
to understand the quality of the sampling. If the traces appear quite randomic,
as they are in this figure, there is nothing to worry. Otherwise
we have to increase the length of the chain so that it can visit
each `point' (in fact a little volume)
of the space of possibilities with relative frequencies `approximately equal'
to their probabilities (just Bernoulli theorem, nothing
to do with the `frequentist definition of probability').
, drawn
automatically by R when
the command plot() is called with first argument an MCMC
chain object, shows, for each of the three variables that we have
chosen to monitor,
the `trace' and the 'density'.
The latter is a smoothed representation of the histogram
of the possible occurrences of a variable in the chain.
The former shows the `history'
of a variable during the sampling, and it is important
to understand the quality of the sampling. If the traces appear quite randomic,
as they are in this figure, there is nothing to worry. Otherwise
we have to increase the length of the chain so that it can visit
each `point' (in fact a little volume)
of the space of possibilities with relative frequencies `approximately equal'
to their probabilities (just Bernoulli theorem, nothing
to do with the `frequentist definition of probability').
Here is the relevant output of the script:
1. Empirical mean and standard deviation for each variable,
plus standard error of the mean:
Mean SD Naive SE Time-series SE
r1 1.334 0.6671 0.002110 0.002110
r2 1.167 0.4418 0.001397 0.001397
rho 1.333 0.9444 0.002986 0.002986
2. Quantiles for each variable:
2.5% 25% 50% 75% 97.5%
r1 0.3637 0.8457 1.223 1.700 2.927
r2 0.4701 0.8481 1.111 1.424 2.179
rho 0.2772 0.7048 1.102 1.684 3.770
Exact:
r1 = 1.333 +- 0.667
r2 = 1.167 +- 0.441
rho = 1.333 +- 0.943
As we can see, the agreement between the MCMC and the exact results,
evaluated from Eqs. (![[*]](crossref.png) )
and (
)
and (![[*]](crossref.png) ),
is excellent (remember that `r1 = 1.333 +- 0.667'
stands for
E
),
is excellent (remember that `r1 = 1.333 +- 0.667'
stands for
E